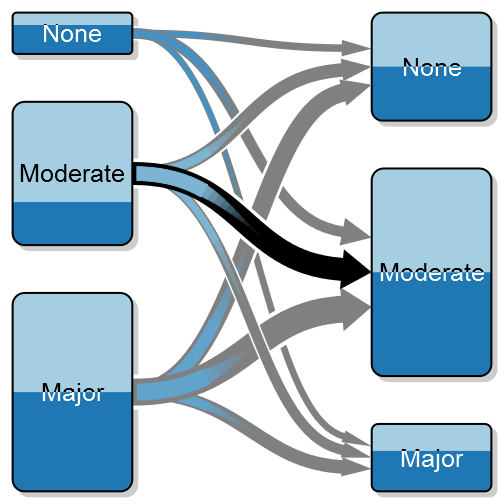
A transition between states – the above is a simulation of before and after surgery where I’ve highlighted the large proportion that doesn’t improve in the moderate group.
As an orthopaedic surgeon I’m often interested in how a patient is doing after surgery compared to before. I call this as a transition between states, e.g. severe pain to moderate pain, and in order to better illustrate these transitions I’ve created something that I call a transition plot. It’s closely related to the plotMat for plotting networks but aimed at less complex relations with only a one-way relation between two groups of states.
This project started by me posting a question on Stack Overflow, the answers were (as always) excellent, but didn’t really satisfy my needs. What I wanted was a graphically appealing plot that I could control in extreme detail. Thanks to Paul Murrell’s excellent grid package I was able to generate a truly customizeable transition plot.
In this post I’ll give a short introduction with examples to what you can do with the transitionPlot()-function. I’ll try to walk you through simple transitions to more complex ones with group proportions and highlighted arrows.
Simulate some data
To illustrate a simple example we’ll generate some data, imagine having a questionnaire for pre- and postoperative pain graded none, moderate, or major. We have the two variables in our data set, and to see the relation we just do a three-by-three table:
library(Gmisc)
library(Gmisc)
# Generate some fake data
set.seed(9730)
b4 <- sample(1:3, replace = TRUE, size = 500, prob = c(0.1, 0.4, 0.5))
after <- sample(1:3, replace = TRUE, size = 500, prob = c(0.3, 0.5, 0.2))
b4 <- factor(b4, labels = c("None", "Moderate", "Major"))
after <- factor(after, labels = c("None", "Moderate", "Major"))
# Create the transition matrix
transition_mtrx <- table(b4, after)
# Create a table with the transitions
htmlTable(transition_mtrx, title = "Transitions", ctable = TRUE)| Transitions | None | Moderate | Major |
|---|---|---|---|
| None | 13 | 33 | 8 |
| Moderate | 50 | 107 | 30 |
| Major | 78 | 131 | 50 |
Simple example
This is interpretable, but understanding the flow can be challenging and time-consuming. Visualizing it using the transition plot gives a quick and more intuitive understanding:
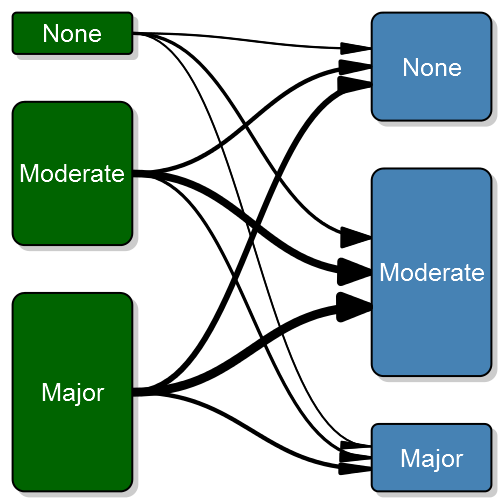
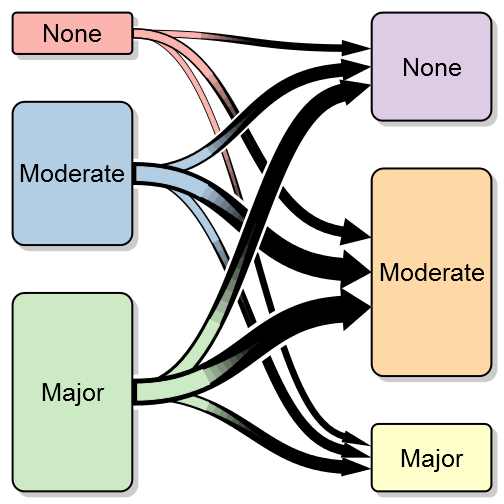
transitionPlot(transition_mtrx)Here you can see the thick lines representing the transition from that particular group into the next. So far I've used the graph for the same measure on left and right side but I guess your imagination sets the limit. It can just as well be two or more treatment arms with some discrete outcomes.
Simple with a twist
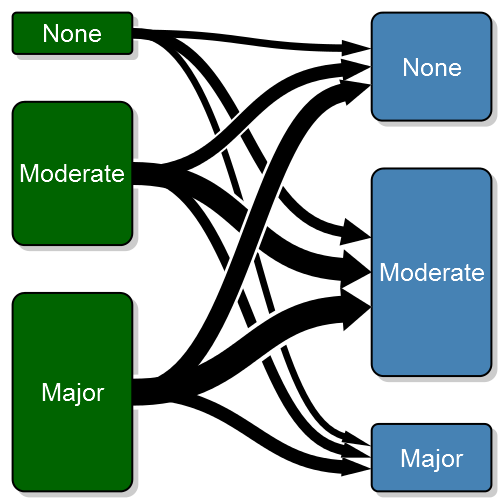
In my previous post I documented how I created an arrow that allows perfect control over start, stop and width. Another annoying detail in the default bezier line, is that the arrows are perpendicular to each box, thus not following the line contour as expected. If we change the arrow type to my alternative arrow we get a nicer plot:
# Note that when using my arrow type you need to specify exact unit width
# since I wasn't certain how the lwd should be translated.
transitionPlot(transition_mtrx, overlap_add_width = 1.3, type_of_arrow = "simple",
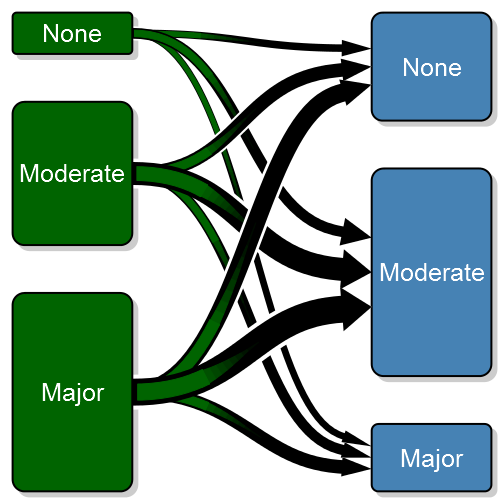
min_lwd = unit(2, "mm"), max_lwd = unit(10, "mm"))Note that I've also added a white background to each arrow in order to visually separate the arrows (the overlay order can be customized with the overlap_order parameter). To further enhance the transition feeling I have added the option of a color gradient. To do this you need to specify arrow_type = “gradient”:
# Note that when using my arrow type you need to specify exact unit width
# since I wasn't certain how the lwd should be translated.
transitionPlot(transition_mtrx, overlap_add_width = 1.3, type_of_arrow = "gradient",
min_lwd = unit(2, "mm"), max_lwd = unit(10, "mm"))More complex coloring
Additionally you can choose different colors for your boxes:
library(RColorBrewer)
transitionPlot(transition_mtrx, txt_start_clr = "black", txt_end_clr = "black",
fill_start_box = brewer.pal(n = 3, name = "Pastel1"),
fill_end_box = brewer.pal(n = 6, name = "Pastel1")[4:6],
overlap_add_width = 1.3, type_of_arrow = "gradient",
min_lwd = unit(2, "mm"), max_lwd = unit(10, "mm"))Split boxes into proportions
Sometimes you might want to split the color of each box into two colors, illustrating a proportion, for instance if we do wrist fracture surgery it might be interesting to explore if the left:right ratio differs. Note that the gradient color is a blend between the two proportions:
transitionPlot(transition_mtrx,
box_prop = cbind(c(0.3, 0.7, 0.5), c(0.5, 0.5, 0.4)),
txt_start_clr = c("black", "white"), txt_end_clr = c("black", "white"),
fill_start_box = brewer.pal(n = 3, name = "Paired")[1:2],
fill_end_box = brewer.pal(n = 3, name = "Paired")[1:2],
overlap_add_width = 1.3, type_of_arrow = "gradient",
min_lwd = unit(2, "mm"), max_lwd = unit(10, "mm"))Highlighting certain arrows
Another interesting option may be to highlight a certain transition:
arr_clrs <- c(rep(grey(0.5), times = 3),
c(grey(0.5), "black", grey(0.5)),
rep(grey(0.5), times = 3))
transitionPlot(transition_mtrx, arrow_clr = arr_clrs,
overlap_order = c(1, 3, 2),
box_prop = cbind(c(0.3, 0.7, 0.5), c(0.5, 0.5, 0.4)),
txt_start_clr = c("black", "white"), txt_end_clr = c("black", "white"),
fill_start_box = brewer.pal(n = 3, name = "Paired")[1:2],
fill_end_box = brewer.pal(n = 3, name = "Paired")[1:2],
overlap_add_width = 1.3, type_of_arrow = "gradient", min_lwd = unit(2, "mm"),
max_lwd = unit(10, "mm"))Summary
Even though it may seem like a very simple plot, after some tweaking I believe the data-per-ink ratio turns out quite OK. I hope you'll find it as useful as I have. As always, don't hesitate to comment. The function transitionPlot is included in my Gmisc-package.

This is absolutely fantastic!
I was able to download the Gmisc package, but transitionPlot() was not a part of it (?) looks neat though. 🙂
Was able to follow the stackoverflow code, neat stuff, thank you.
http://stackoverflow.com/questions/16112162/graph-flow-chart-of-transition-from-states
Are you using R ver. 2.15? The Gmisc-package version 0.5.3.3 available for 3.0 has the function. I guess installing with the source option should work just as well, alternatively you can find the most recent code on GitHub: github.com/gforge/Gmisc
I was just playing with this plot again and realized I couldn’t control some finer points. Specifically, adding a title or combining multiple plots, e.g., par(mfrow=c(2,2)) or multiplot() http://www.cookbook-r.com/Graphs/Multiple_graphs_on_one_page_(ggplot2)/
I thought you might point me in a direction if you had a moment. Would it require some study of the grid package?
I’ve updated the package so that it doesn’t call plot.new(), that is now you can use it inside you current viewport, i.e. just set your title in one viewport and then use another one for the actual plot. The same thing goes for multiple plots.
Great, thanks. your older version on stack overflow wasn’t nearly as refined. Neat graphic, bet it will prove useful.
Great idea!
I’m just about to give this a whirl, daft question though – if I wanted to see these transitions over multiple points in time like Before-After-1 year later, would I need to build 2 seperate matrices, two charts and overlay them somehow?
Hi Steph,
Yes, currently you would need to overlay two plots. I figured that it would be too clotted with more than two states. If you’re interested you can just modify the code a little to get it to work with multiple layers, you need to adapt the getBoxPositions function to make it more general and also rewrite some of the logic to allow multiple transition matrices. There are a lot of customizeable details that will take some thinking in order to make them more general and still easy to use.
I’ve gone for an easier time-series approach instead and chucked my categories on the y axis and my snapshot points on the x axis, using a low alpha so that the stronger the colour the more travelled the path.
Still, awesome chart – thank you!
Pingback: Using SVG graphics in blog posts – Mubashir Qasim